Class I major histocompatibility complex (MHC-I) proteins are crucial in the immune system of all jawed vertebrates. They showcase peptide fragments of proteins present inside the cell on the cell surface, displaying them to the immune system. This allows the immune system to scan the body for foreign or toxic antigens. When foreign peptides are detected, they trigger a cascade that enables cytotoxic T cells to eliminate intruders. This process has been used for the development of vaccines and immunotherapy. Researchers identify fragments of peptides unique to viruses or cancer, screen for T cells that detect those targets, and initiate an immune response. However, the current method of using MHC-I molecules as probes in vaccine and immunotherapy development is time-consuming and inefficient.
Stabilizing MHC-I Molecules for Rapid Production
Researchers from Children’s Hospital of Philadelphia (CHOP) have developed stable, universal MHC-I molecules that can be produced rapidly at scale. This allows researchers to develop vaccines and immunotherapies more quickly and identify molecules that can work broadly across the population. The findings of the research were published in the Proceedings of the National Academy of Sciences.
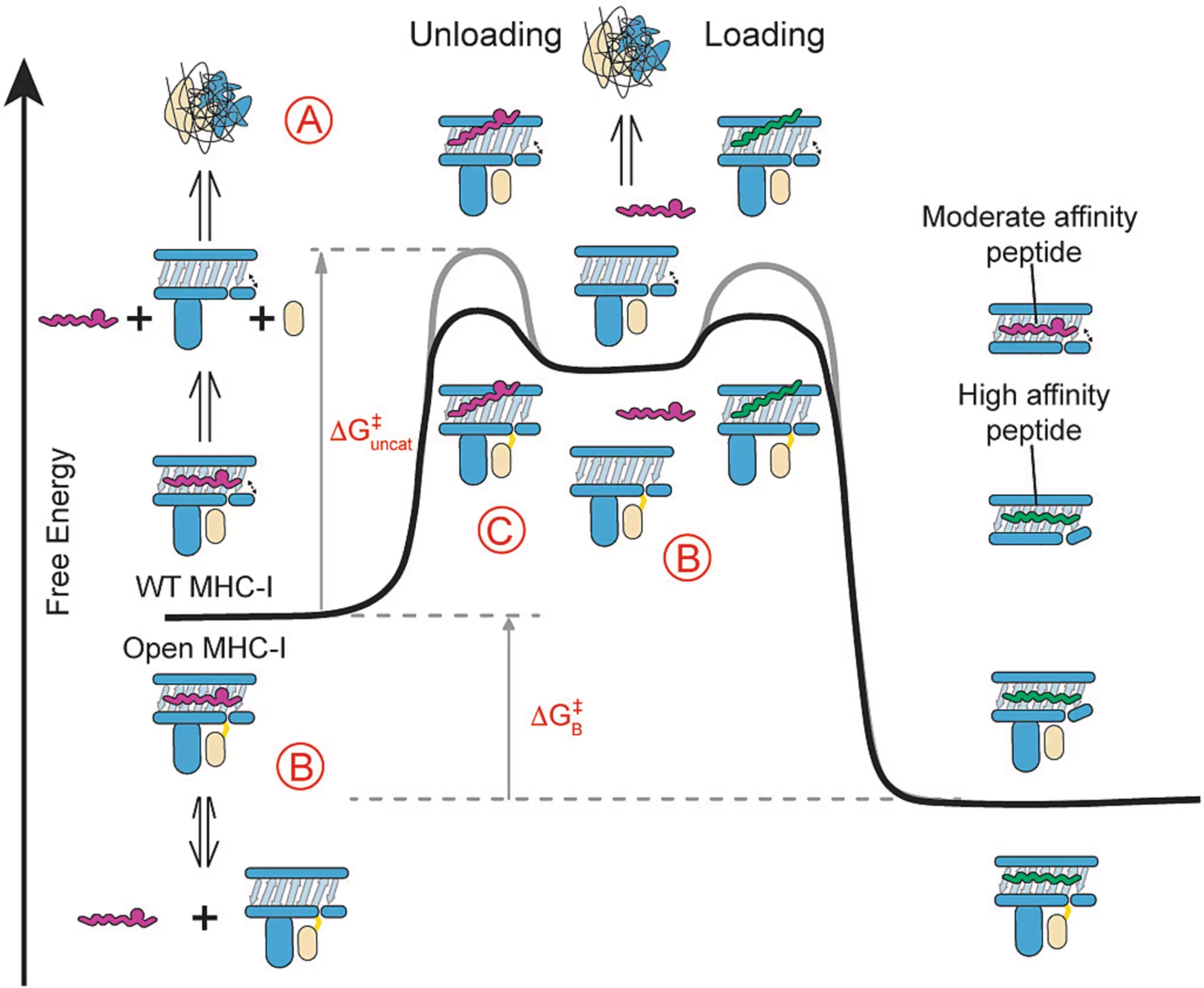
Led by Sgourakis Lab members Yi Sun, Michael C. Young, and Claire H. Woodward, the researchers stabilized MHC-I molecules by focusing on their 3D structure. They focused on the light chain of the molecule since it is conserved across HLA types. The light chain plays a vital role in stabilizing MHC-I molecules. As soon as the light chain falls off, it triggers the MHC-I molecule to disassemble, at which point it is sent for recycling for future peptide loading. To exploit this fundamental engineering, the researchers tethered the heavy chain to the light chain, stabilizing the MHC-I in a conformation that is “open” and receptive to peptides.
Confirmation of Stability and Enhanced Peptide Exchange
The researchers confirmed the stability of these engineered MHC-I molecules through biophysical characterization using nuclear magnetic resonance (NMR). They demonstrated that their open MHC-I molecules have enhanced stability when loaded with peptides, even those of low to moderate affinity. The researchers also showed that their engineered MHC-I molecules promote peptide exchange across multiple HLA allotypes, covering representatives from five HLA-A, six HLA-B supertypes, and HLA-Ib molecules, which have many different forms.
A high-throughput peptide exchange platform was enabled by the design of customized “placeholder” peptides containing unnatural amino acid modifications. This design was done in close collaboration with the lab of chemical biologist George Burslem, Ph.D., at the University of Pennsylvania.
Potential for Revolutionizing Vaccine Development
The findings in this paper have the potential to revolutionize the field of vaccine development and the rate at which researchers can screen for effective and universal immunotherapies. According to senior author Nikolaos G. Sgourakis, Ph.D., Associate Professor in the Center for Computational and Genomic Medicine at Children’s Hospital of Philadelphia, these stabilized MHC-I molecules could speed up the screening process and the subsequent development of effective therapies.
Dr. Sgourakis added that the open MHC-I platform leverages minimal protein modifications to enhance ligand exchange reactions across all known HLA allotypes. The new molecules could be a versatile tool for screening antigenic epitopes, enabling the detection of low-frequency receptors and engineered antibodies for the development of targeted therapies.


Leave a Reply