The COVID-19 pandemic has wreaked havoc across the globe, leading to widespread sickness and multiple national lockdowns. While certain countries are starting to experience a decline in infection rates, cases still persist, emphasizing the need for a deeper understanding of the mechanisms that sustain the virus within the body. Researchers at Kanazawa University have delved into the role of the open reading frame 6 (ORF6) protein in COVID-19 symptoms, hoping to uncover new insights that could aid in the development of more effective treatments and prevention strategies against this devastating disease.
For a virus like SARS-CoV-2 to successfully replicate and survive, it produces various accessory proteins that reprogram the host environment in its favor. ORF6 is one such accessory protein that has attracted significant attention due to its potential role in interfering with the function of interferon 1 (IFN-I) – a crucial component of the immune system. Evidence suggests that ORF6 may be responsible for the retention of specific proteins in the cytoplasm while disrupting mRNA transport, thereby inhibiting IFN-I signaling and potentially leading to asymptomatic infections.
Unraveling the Structure of ORF6
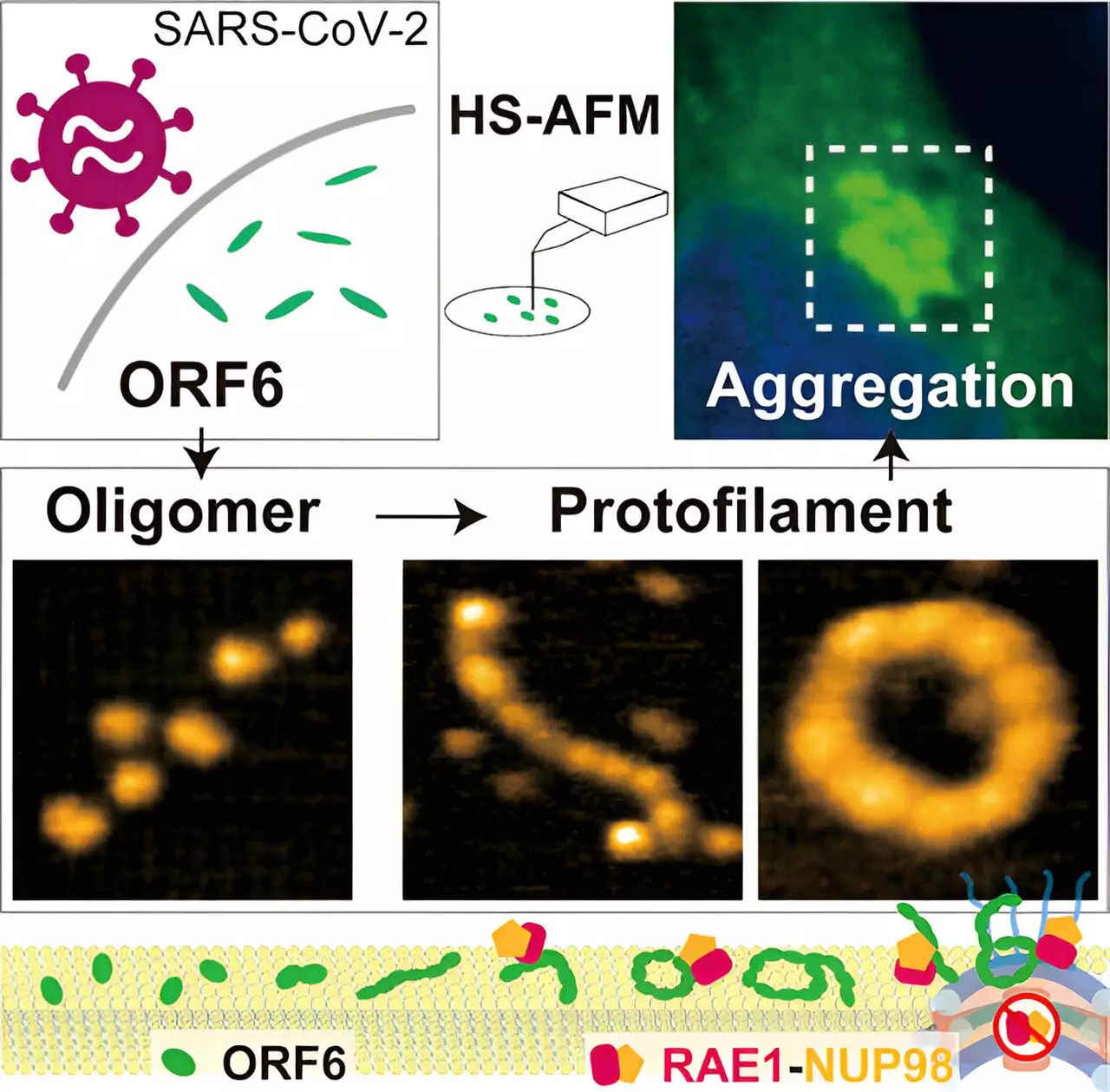
To shed light on the mechanism behind ORF6’s actions, the researchers employed various software programs and nuclear magnetic resonance measurements to analyze its structure. Intrinsically disordered regions were identified, limiting the use of machine learning algorithms like AlphaFold2. Instead, high-speed atomic force microscopy (HS-AFM) was utilized, enabling the researchers to examine the topography of ORF6 samples at a high resolution. Through HS-AFM, it was discovered that ORF6 primarily forms ellipsoidal filaments of oligomers, which are shorter than polymers. Interestingly, these filaments exhibited variations in length and circumference depending on temperature and the presence of lipids.
HS-AFM not only revealed the structures of ORF6 but also provided insights into its behavior. The researchers observed circular motion, protein assembly, and flipping, offering a glimpse into the dynamics of ORF6. Furthermore, computer analysis indicated that these filaments had a propensity to aggregate into amyloids, similar to those found in some neurodegenerative diseases. Such aggregation could result in complications in COVID-19 symptoms, as it effectively sequesters numerous host proteins, especially transcription factors involved in IFN-I signaling.
Understanding the way ORF6 functions opens the door to potential therapeutic interventions. The researchers found that the filaments were predominantly held together by hydrophobic interactions. Consequently, they suggest exploring druggable candidates that can disrupt these interactions and dissociate ORF6 aggregates. Compounds such as certain alcohols, urea, or sodium dodecyl sulfate were shown to break up the filaments, indicating possible avenues for therapeutic intervention. The evaluation of these candidates and their therapeutic value in tackling COVID-19 requires further investigation.
Through their high-speed atomic force microscopy studies, researchers at Kanazawa University have shed light on the structure and behavior of ORF6, an accessory protein in SARS-CoV-2. Understanding the role of ORF6 in COVID-19 symptoms provides valuable insights into potential treatments and prevention strategies. The identification of hydrophobic interactions as a key factor in maintaining the integrity of the protein opens up avenues for developing targeted therapeutics that can disrupt these interactions and mitigate the detrimental effects of ORF6. Further research in this area will contribute to the ongoing battle against COVID-19, and potentially pave the way for future outbreak preparedness.


Leave a Reply