Protein synthesis is a fundamental process in cells, and ribosomes play a vital role as the cell’s “protein factories.” The ribosomes are responsible for converting genetic information into proteins, which are essential for the functioning of living organisms. While the assembly of ribosomes has been extensively studied, the degradation of ribosomes has remained relatively unexplored. In a recent study published in the scientific journal Nature, a research team from the Department of Chemistry at the Universität Hamburg has successfully identified the molecular mechanism employed by the enzyme RNase R in degrading the ribosomal 30S subunit.
Under stress situations like nutrient scarcity or during the stationary phase of cell growth, cells reduce their metabolic activities to enhance their survival. In this state, energy-consuming processes like protein synthesis are decreased, and some ribosomes are degraded to release the invested energy. However, the structural details of ribosomal degradation have been lacking, making it a critical area of investigation.
The researchers chose to study Bacillus subtilis, a common soil bacterium found in various environments. Unlike previous studies conducted during the stationary phase, the researchers focused on cells that were still actively growing. This enabled them to understand the processes occurring during the transition to the stationary phase more effectively.
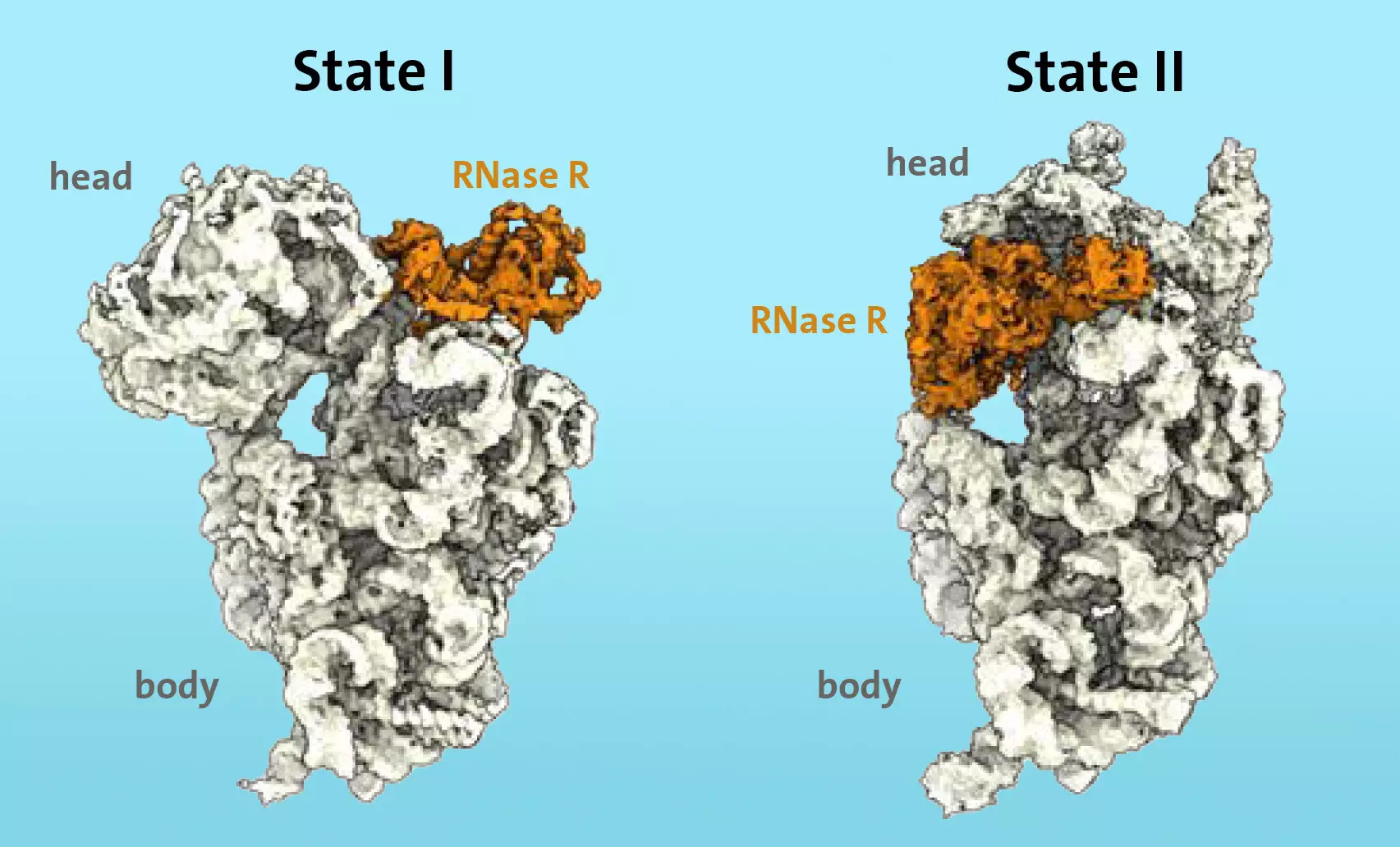
Using cryo-electron microscopy, the research team observed the binding of the enzyme RNase R to the small 30S subunit of the ribosome. The researchers discovered that RNase R binds specifically to a region called the “neck” of the ribosomal subunit. They then demonstrated that RNase R destabilizes the neck area in two stages, effectively detaching the “head” of the subunit. This detachment allows RNase R to continue the degradation process of the 30S subunit unimpeded.
The researchers’ in-vitro degradation experiments confirmed that RNase R alone is sufficient to complete the degradation process of the 30S subunit. This finding highlights the significant role of RNase R and its enzymatic activity in ribosomal degradation. Additionally, the study provides valuable insights into the kinetic barriers that RNase R overcomes during the degradation process.
The knowledge gained from this research contributes to a comprehensive understanding of cellular processes during stress and the stationary phase. By uncovering the molecular mechanism of ribosomal degradation, scientists can further investigate its regulation and potential therapeutic interventions. Targeting the degradation mechanism could be a novel approach to modulate ribosomal turnover and protein synthesis, which has implications in various biological and pathological scenarios.
The groundbreaking research conducted by the team from the Universität Hamburg’s Department of Chemistry provides significant insights into the molecular mechanism of RNase R in degrading the ribosomal 30S subunit. The study sheds light on the dynamic process of ribosomal degradation during stress and offers a promising avenue for future investigations. Continuing to explore the intricate details of this process will deepen our understanding of cellular mechanisms and potentially open new therapeutic avenues.


Leave a Reply