Lipids are vital biomolecules that play a crucial role in numerous cellular processes. To gain a comprehensive understanding of these processes, it is necessary to analyze and characterize all the lipids present in a sample. This field of study, known as lipidomics, provides valuable insights into complex biological systems. However, a major challenge in lipidomics research lies in connecting the diverse structures of lipids to their specific biological functions.
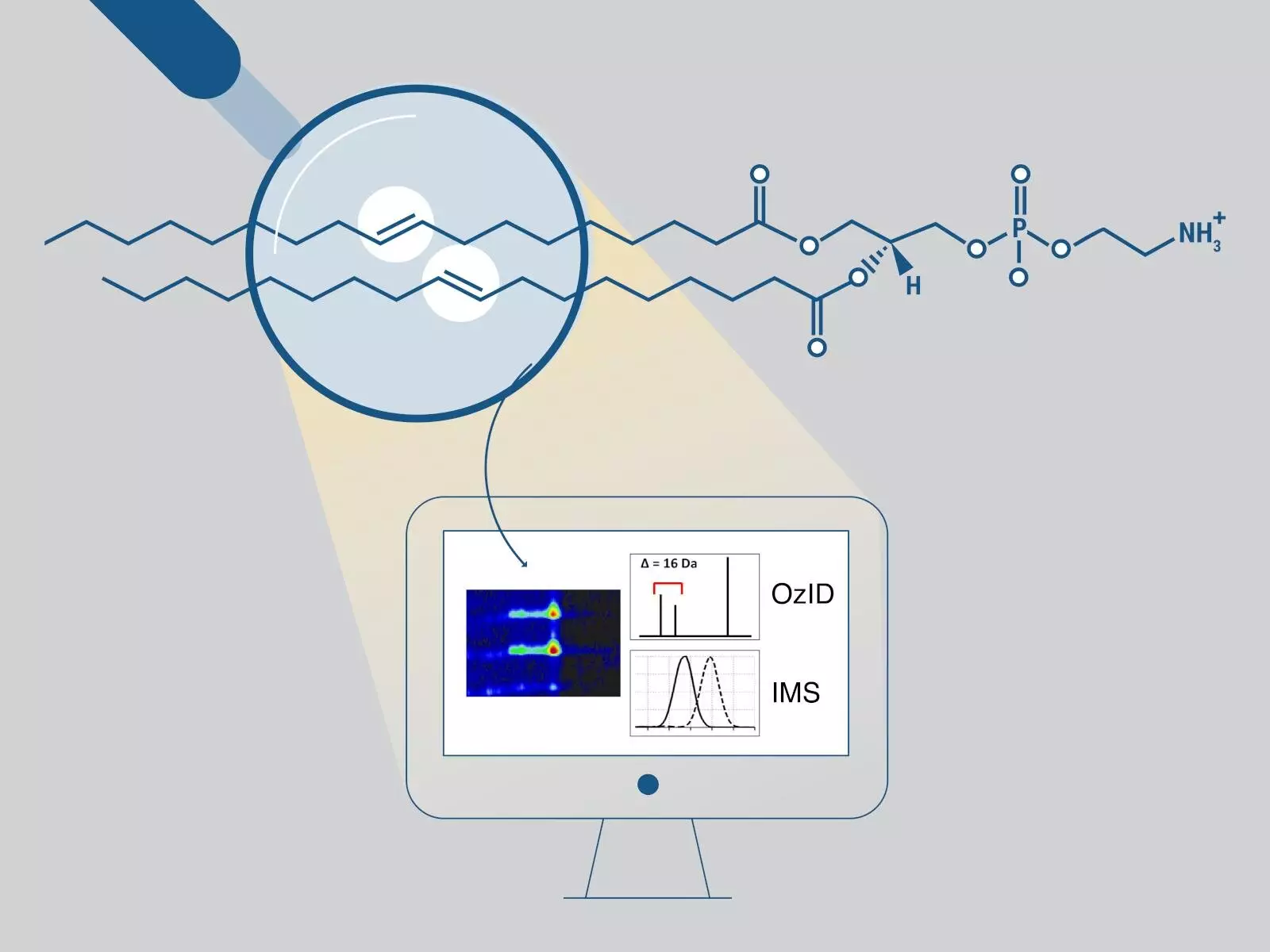
Among the various characteristics of lipids, the positions of double bonds within fatty acid chains have garnered significant attention. This is because these double bonds can influence the physical properties of cellular membranes and modulate cell signaling pathways. Unfortunately, determining the exact positions of double bonds is not a routine practice in lipidomics studies. The experimental setup required to achieve this is complex, and the resulting data is often intricate.
To address this challenge, scientists at Pacific Northwest National Laboratory (PNNL) have developed a revolutionary workflow that combines automation and machine learning approaches. Their innovative method, known as LipidOz, significantly streamlines the analysis of lipid data to determine the positions of double bonds accurately.
LipidOz represents a significant advancement in lipid characterization, offering researchers a more efficient and precise method. Its development caters to the unique requirements of lipidomics studies and fills a crucial gap in the current analytical tools available.
The identification of lipids poses a challenge due to the existence of molecular components with the same chemical formula but varying physical configurations. These components differ in attributes such as fatty acyl chain length, stereospecifically numbered (sn) position, and the position/stereochemistry of double bonds.
Conventional analyses can determine fatty acyl chain lengths, the number of double bonds, and, in some cases, the sn position. However, they fall short when it comes to determining the positions of carbon-carbon double bonds. Overcoming this limitation requires employing the gas-phase oxidation reaction known as ozone-induced dissociation (OzID), which generates distinctive fragments aiding in double bond position determination.
The complex and repetitive analysis of the data obtained from OzID poses another challenge. Additionally, the lack of software tools specifically designed for this purpose further hampers progress in the field. To address these issues, PNNL researchers have created LipidOz, an open-source Python tool that automates the process of determining and assigning double bond positions in lipids.
LipidOz utilizes a combination of traditional automation techniques and deep learning approaches to accurately identify and assign these crucial double bond positions. The tool has been successfully tested on both standard lipid mixtures and complex lipid extracts, demonstrating its practical application in future lipidomics investigations.
LipidOz represents a significant breakthrough in the field of lipidomics, offering researchers a more efficient and accurate method for determining double bond positions within lipids. By streamlining the data analysis process, LipidOz eliminates the complexities associated with this task. This innovative tool opens up new avenues for exploring the connection between lipid structures and their biological functions. As the field of lipidomics continues to advance, LipidOz provides a vital resource for researchers seeking to unravel the intricate mechanisms underlying cellular processes.


Leave a Reply