Researchers at Cornell University have developed a new method for analyzing protein crystals that could revolutionize drug discovery. The method, which was outlined in a paper published in Nature Communications, allows researchers to interpret the data from X-ray crystallography experiments that was previously discarded. This development could lead to a better understanding of a protein’s movement, structure, and overall function.
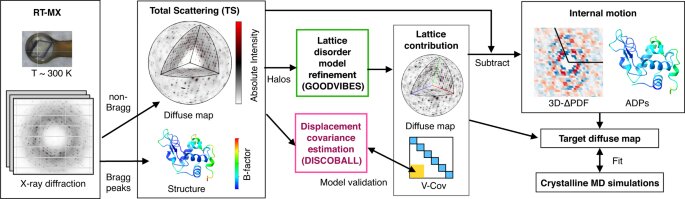
Protein crystallography produces bright spots known as Bragg peaks from the crystals, which provide high-resolution information about the shape and structure of a protein. This process also captures blurry images, patterns, and clouds related to the movement and vibrations of the proteins hidden in the background of the Bragg peaks. These background images are typically discarded, with priority given to the bright Bragg peak imagery that is more easily analyzed.
The Two-Part Method
The researchers developed a two-part method called GOODVIBES and DISCOBALL, which simultaneously provides a high-resolution structure of the protein and information on its correlated atomic movements. GOODVIBES analyzes the X-ray data by separating the movements of the protein from other proteins that might be moving around it. DISCOBALL independently validates these movements for certain proteins directly from the data, allowing researchers to trust the results from GOODVIBES and understand what the protein might be doing.
According to lead author Steve Meisburger, the information related to the motion of the atoms of the protein is there, but researchers didn’t know how to use it. Meisburger worked closely with associate professor of chemistry and chemical biology Nozomi Ando to develop the robust workflow to decode the weak background signals from crystallography experiments called diffuse scattering. This allows researchers to analyze the total scattering from crystals, which depends on both the protein’s structure and the subtle blur of its movements.
Ando said that while the potential for using diffuse scattering has been recognized for a long time, accurately measuring the subtle signal while processing the data for something useful has been very difficult to do. However, she believes that turning GOODVIBES and DISCOBALL into a genuine structural technique that can be used by researchers at synchrotrons all over the world will be a game-changer.
In conclusion, the development of this method could open up applications for new drug discovery and other areas of biotechnology and biochemistry. By isolating the internal motion signals from total scattering data of complex proteins, researchers can learn more about how proteins move and interact with other important molecules, which can be used to design new drugs and therapies that target specific proteins.

Leave a Reply